By: Louisa Gaylord
The majority of mental health disorders are first diagnosed in childhood or adolescence. The disorders most commonly detected in young people include ADHD, anxiety, depression and behavior problems. Historically, individual labs and researchers interested in understanding the brain basis of mental health only had access to small amounts of data to analyze, like MRI scans, and for many procedures the data needed to be examined in detail by a professional. But as data science approaches have become more prevalent in neuroscience and other fields, researchers now have access to data collected from thousands of individuals. The sheer amount and complexity of these new datasets have made it difficult to scale up the historic approach.
“In the last few years, large collaborative research projects are openly sharing data and [neuroscience] is gaining access to unprecedented amounts of information about the structure and function of the brain in various states of health and disease,” said Ariel Rokem, Research Assistant Professor of Psychology and eScience Data Science Fellow at the University of Washington. “But the size of these datasets and their complexity also pose significant barriers to research; comprehensive analysis requires a large amount of computational power.”
In a nuanced process like analyzing MRI scans, how can additional computational power be added in a way that the intricate data is preserved? Rokem and UW Data Science Postdoctoral Fellow Adam Richie-Halford have created Fibr, a way for citizen scientists – that is, anyone who wants to participate – to “teach” quality control to an algorithm that is learning to read child and adolescent MRI scans.
The initial framework of Fibr is based on Braindr, a similar project developed several years earlier by eScience Institute Postdoctoral Fellow Anisha Keshavan and her co-mentors, Rokem and Jason Yeatman with UW’s Institute for Learning and Brain Sciences. When Adam Richie-Halford joined the team as a Postdoctoral Fellow in the summer of 2020, he and Rokem realized that they could get even more out of the project: “The Braindr platform could be extended to review diffusion MRI (dMRI) data, not just the structural MRI data used in Braindr,” said Richie-Halford.
The dMRI measures long-range fiber connections between different brain regions. “Our algorithms map the connections between brain regions using open-source software we developed,” Rokem says – the Diffusion MRI in Python (DIPY) project, which was awarded a three-year NIH grant in 2018. The following year, an additional grant was awarded from NIH to Rokem, Yeatman and Noah Simon, their collaborator in the UW Department of Biostatistics, for a project that focused specifically on algorithms to analyze large shared datasets, such as the Human Connectome Project. By tracking these connections across thousands of patients with developing brains, researchers can learn more about how they play a role in mental health disorders.
Fibr is created around a similarly large dataset, the Healthy Brain Network (HBN), a pediatric mental health study by the Child Mind Institute (CMI) that has been collecting MRI scans of the brains of children ages 5-20 in the hopes of learning more about mental health disorders. Before the team could analyze the data, they realized that the raw dMRI data would need to be preprocessed. “dMRI is a promising and rich modality but the raw data requires preprocessing to correct for head motion and other issues,” said Rokem. They coordinated with members of the CMI research team and the Lifespan Informatics & Neuroimaging Center (LINC) with director Ted Satterthwaite at the University of Pennsylvania, to agree on a community standard for the preprocessing pipeline.

To begin with, the Child Mind Institute collects and anonymizes the child and adolescent MRI dataset provided by the Healthy Brain Network. The dMRI images are treated differently than the participants’ health and demographic info. Each participant is assigned an anonymized participant ID and their personal details, such as their socio-economic status and psychological assessment, are inaccessible to researchers unless they have signed a strict data usage agreement. “Each MRI image is assigned an anonymized participant ID and the participant’s face is removed from the images,” explained Rokem. “After that, the images are made publicly available through releases by the International Neuroimaging Data-Sharing Initiative and 1000 Functional Connectomes Project.”
With thousands of scans to analyze, the individual attention that works on a small scale becomes impractical on a much larger one. In order to train a computer algorithm to correctly identify poor quality data or failures in the preprocessing pipeline, “we need labelled data to train the quality control algorithms,” said Richie-Halford. “The preprocessing pipeline is automated through a software developed by Matt Cieslak at Penn LINC, but experts still need to perform quality control on the end results to make sure that it’s performing as expected.”
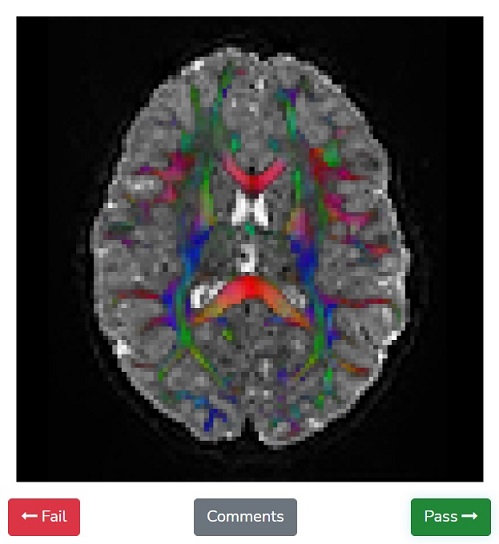
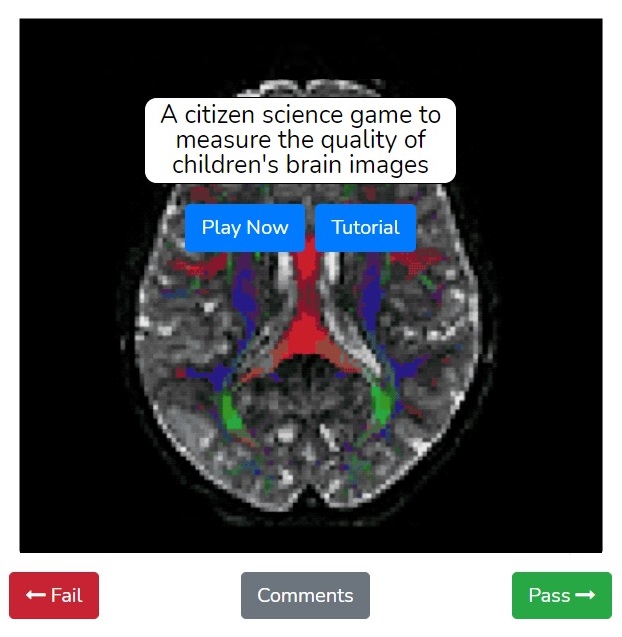
When you log into the Fibr website, you have the opportunity to view the anonymized MRI data and swipe left or right, depending on whether the MRI scan displayed shows the right pattern of long-range fiber connections. This public input provides the training data that helps guide the quality control algorithm to identify good or bad dMRI data. Rokem and Richie-Halford have formatted Fibr as a game with a simple tutorial so that anyone can participate, regardless of scientific training or previous experience.
Play Fibr now and join the rankings!

While Fibr and its predecessor Braindr seem like they are only relevant to the neuroscience community, the SwipesForScience platform they are built on is a web app where users can provide feedback on many different kinds of scientific data. “Both the feedback and the data can take different forms,” said Rokem. “The data can be images, animated gifs or audio recordings. And the feedback can vary from pass/fail, a rating scale of 1 to 5, or asking users to draw on the images provided.” The flexibility of SwipesForScience means that it is already being implemented in other fields like pharmacology and cetology.
Additionally, because huge datasets like the dMRI images are relatively new, the field lacks a consensus on the best way to quality control such large data, and the concept of using citizen science in the process is still largely untested. “If Fibr is successful for the dMRI modality, then perhaps future large-scale MRI datasets will be quality controlled by a union of citizen scientists and machines,” Richie-Halford said.
Another beneficial product of the NIH funding for to the Diffusion MRI in Python project has been Cloudknot, a software library that Richie-Halford and Rokem developed that allows researchers all over the world to easily package their computations and move them to the cloud and scale them up for use with other massive datasets. “One of the goals of our research is to make cloud computing easier for researchers,” said Rokem. “We used this software to process the scans from HBN with AWS cloud computing, and we are providing the processed data openly on AWS, so that other researchers can use it.” Cloudknot is now in use in the UW’s Chemical Engineering Department in the Nance Lab to sort through and analyze the huge datasets generated by their microscopy experiments. Rokem and his research group also continue to explore the potential of cloud computing in collaborative neuroscience through the University of Washington Azure Cloud Credits program.
Fibr demonstrates the new possibilities in our ability to analyze vast amounts of data and track new trends in pediatric neuroscience, but there must be a balance between automation of computational power and human quality control. The more citizen scientists who participate in Fibr and help guide the algorithm to identify fiber connections in dMRI scans, the more accurate the end results will be.
“Help us by playing the quality control game that we designed,” said Richie-Halford. “You see a brain image and you swipe left for a “failing” image that has defects, or right for a “passing” image that looks fine.” Participants are ranked on a leaderboard by how many dMRI scans they have swiped and can earn different badges. “Some of our most avid users have already swiped thousands of images!”