Project Lead: Abigail G. Schindler, acting assistant professor, Psychiatry and Behavioral Services
eScience Liaison: Valentina Staneva
Chronic health conditions (e.g. mental health, pain) are increasing in the US and contribute substantially to decreased quality of life, loss of productivity, and increased financial burden. Indeed, the CDC estimates that over 90% of annual health care expenditures are for people with one or multiple chronic health conditions. Translational research efforts using rodent models can provide much needed insight into underlying mechanisms of chronic health conditions and are needed in order to facilitate the search for therapeutic approaches that can reduce and/or prevent adverse/maladaptive outcomes.
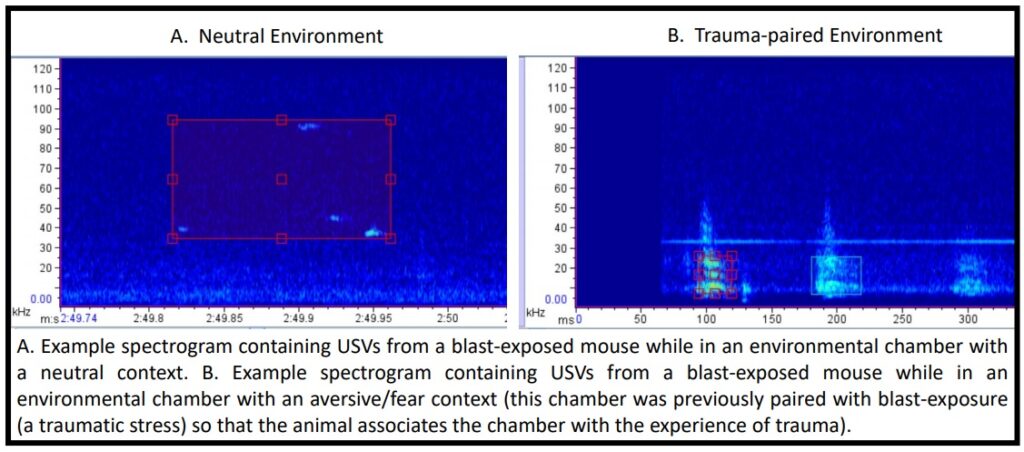
Critically, accurate quantification of affective state (e.g. positive, negative, pain, fear) has historically been a challenge in rodent models, with current available methods suffering from high subjectivity, lack of throughput, and invasive methods, leading to lack of reproducibility across research labs and/or inability to translate to humans. One promising area of research in rodent affective state is ultrasonic vocalizations (USVs). USVs are a form of rodent communication thought to represent an unbiased metric of affective state (there are thought to be potentially different “call signatures” for pleasure, pain, fear, etc.), but are historically difficult to analyze and interpret.
Currently, there is no open-source software available for USV detection and/or analysis (although Matlab based options exist, e.g. DeepSqueak), limiting the applicability of USV research. With a focus on these USVs and open-source products, the current project seeks to develop a Python-based, high-throughput approach for 1) isolating USV calls and 2) assessing affective state. We have USV recordings from a variety of mouse groups (e.g. control, TBI, fear-induction, neuropathic pain) and our goal is to establish specific USV call repertoires/signatures related to specific affective states/experimental conditions/behavioral tasks/therapeutic treatments.
Project Results
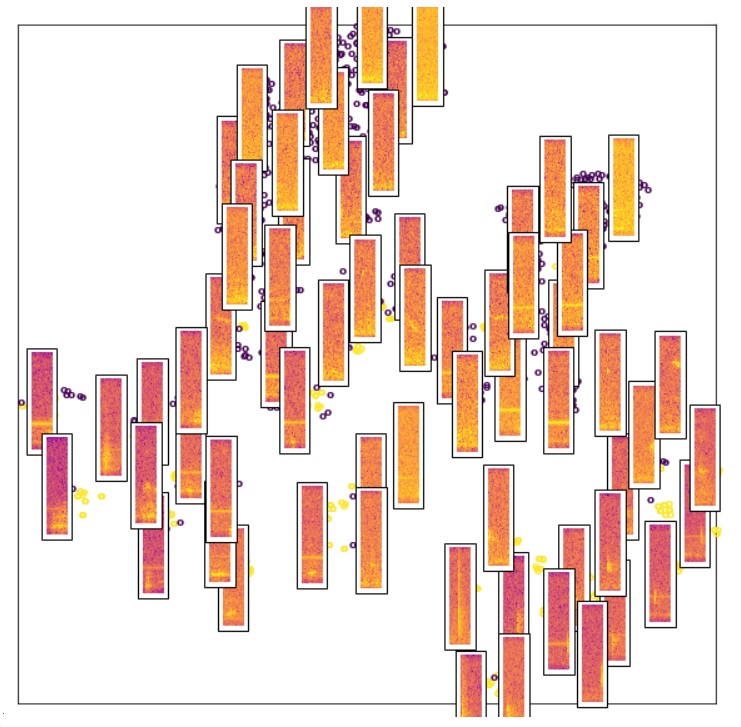
During the incubator we created a series of Python Jupyter Notebooks for processing audio files to isolate USVs using either supervised classification algorithms or transfer learning. Notebooks for visualization and clustering analysis were also created. We utilized Google Colaboratory’s free cloud service with GPU support.
The general data acquisition and analysis pipeline is as follows: 1) acquire audio recording of rodent USVs using Avisoft SASLab Lite (free; saves audio as .wav file), 2) annotate USVs in each file (to use for training classifier) using Raven Lite (free; saves annotations as a ‘selections table’), 3) use annotated selections to train a classification algorithm (USV vs noise), 4) use trained model to process un-annotated audio files.
Audio files were split into 25 ms slices, converted into spectrograms, and saved in a labelled array format for easy access afterwards. Two feature sets were generated and used for model testing and evaluation of shallow learning algorithms: a) set of 8 spectral features (power, purity, centroid, spread, skewness, kurtosis, slope, and roll off), b) power spectrum distribution used as features (e.g. 257 frequencies contained in spectrogram, find power at each frequency for each slice). Full spectrogram images were used for deep learning. The final chosen model was saved and then used subsequently to isolate USVs from un-annotated audio files.
An initial challenge of sufficient computer RAM for processing the audio files was overcome by using the Python package Xarray and saving the processed data (e.g. 25 ms spectrogram slices) as netCDF files. A second challenge was the highly imbalanced nature of the datasets (e.g. ~25000 slices from a 10 min audio recording will contain ~5-50 USVs). We approached the problem by balancing the training dataset through upsampling, downsampling, and stratification procedures, which although it achieved good performance on the training set, resulted in many false detections on new unobserved data. We concluded that downsampling restricted the diversity of the noise during training and we achieved better performance without balancing by appropriately weighting the objective. Cross-validation with rare observations was a challenge since performance scores become sensitive to the training data organization. This was especially visible in deep learning training when data is split into small batches, while the dimensionality of the features is large.