Project Lead: Dr. Nicholas Reder, Pathology
eScience Liaison: Ariel Rokem
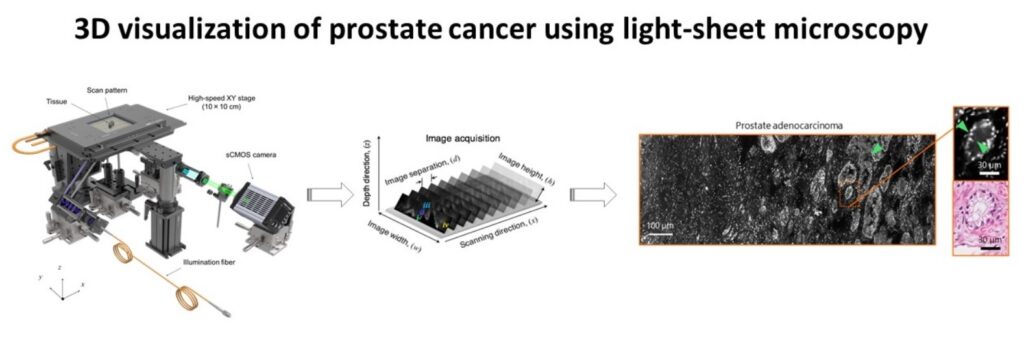
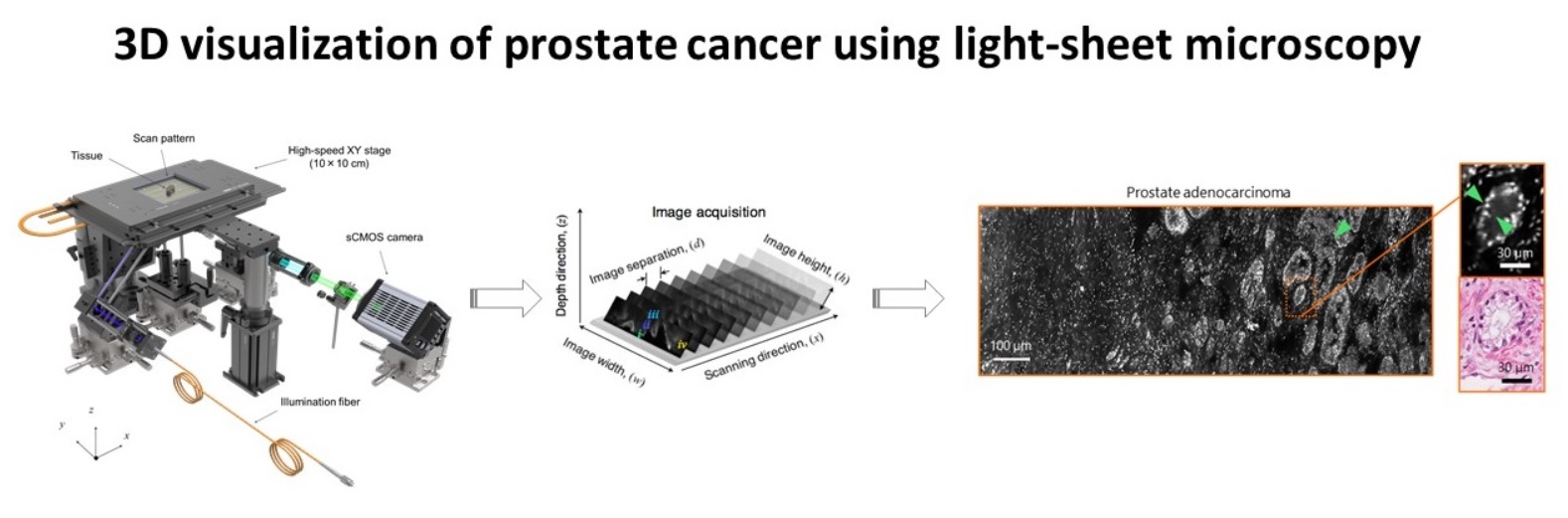
Advances in microscopic imaging enable visualization of heretofore unseen 3D microanatomical features, which have the potential to transform cancer diagnostics. These novel imaging techniques have led to improved diagnostics in kidney biopsies, brain structure, and embryonic development. Light-sheet microscopy and tissue clarification techniques are a particularly intriguing combination because large volumes of cancer tissue can be imaged with high spatial resolution. However, data processing, data management, and visualization of 3D structures has lagged behind the advances in data acquisition. Currently, our data processing steps require 12-24 hours of computing time and are accomplished using fragments of code written in matlab and Miji (matlab + Fiji). In addition, our existing software code is stored on multiple servers and is not well annotated, limiting reproducibility of our work. Finally, the visualization of 3D structures is suboptimal and hinders our ability to provide diagnostic insights. Thus, our current data science limitations have impeded potentially groundbreaking discoveries in cancer microanatomy.
To solve this problem, we will create a software package to optimize the processing, storage, and visualization of 3D microscopic data. Key morphologic features will be extracted from the images and displayed in a simple, easy to navigate format. All code will be well annotated and stored on GitHub to ensure reproducibility.